Project
PI: Dr. Sourav Bhaduri, Assistant Professor
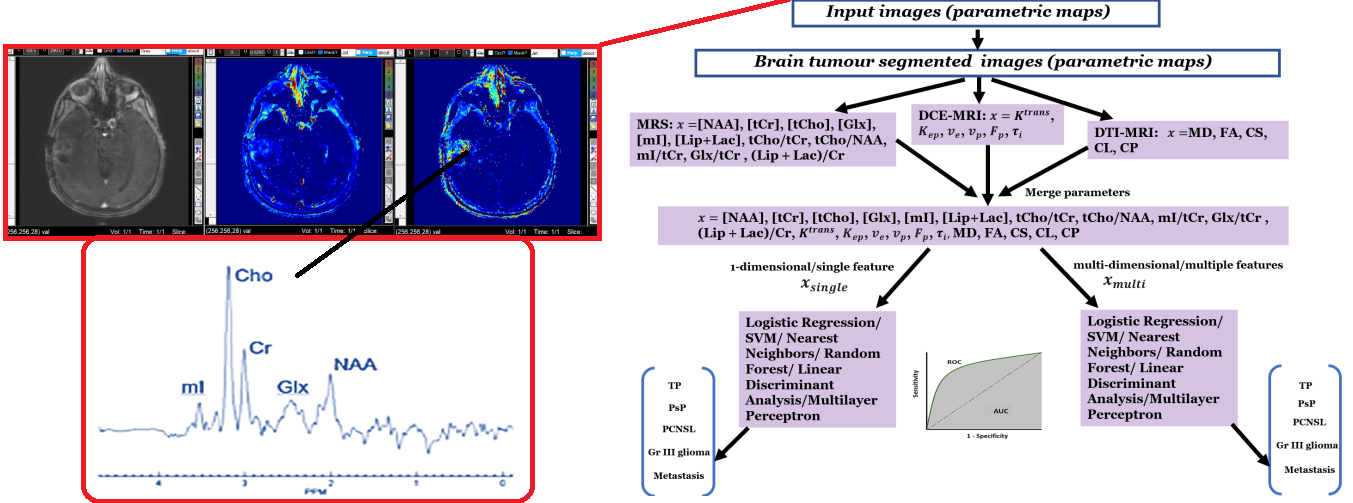
This research develops ‘a. AI/ML and signal/image processing methods for the assessment of brain tumour heterogeneities and treatment response using multi modal MRI parameters and b. Study of Dynamic Contrast Enhanced (DCE) MRI based perfusion Imaging, Diffusion Tensor Imaging (DTI) for measurement of diffusion and high speed/high resolution multi-voxel MR spectroscopic imaging (MRSI) imaging to quantify neurometabolites in different types of brain tumours.’ The project's tasks include collecting data from the above mentioned imaging modalities, develop/implement signal/image processing algorithms, analysing data and developing a GUI toolbox for assessing treatment response in brain tumours. In this research, signal/image processing, advanced MR physics, AI/ML based techniques will be used for medical imaging analysis. MR pulse sequence development will be also be performed to develop/implement faster and accurate methods for data acquisition and advanced data reconstruction algorithms will also be developed/implemented.
We are looking for ambitious young scientists to join this project. The ideal PhD candidate should have : some experience with (medical) signal/image processing, AI/ML, basic knowledge of MR physics and neuroimaging, strong programming skills (MATLAB, or Python) and strong interest in biomedical questions and research.
PI: Dr. Sourav Bhaduri, Assistant Professor
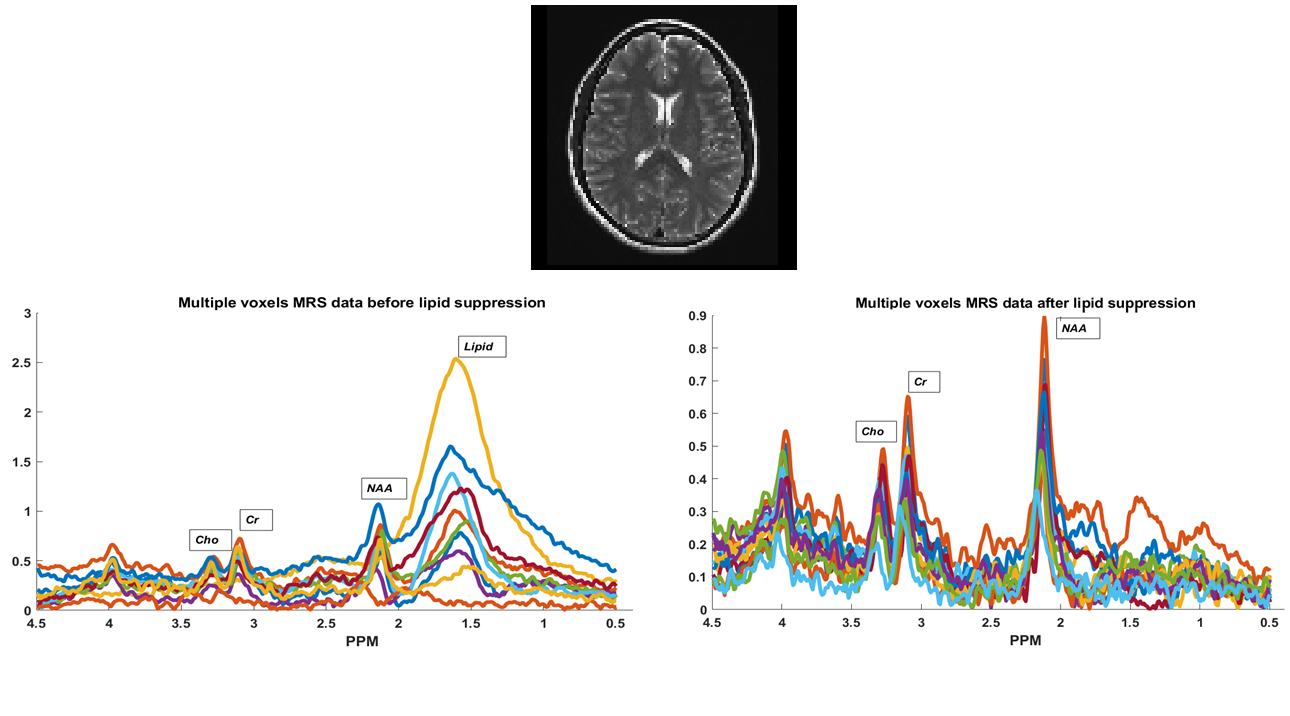
This research develops ‘methods for outer volume brain lipid suppression suppression for preservation and accurate quantification of Lactate peaks (in brain tumour cases) and other main neuro-metabolites in proton MR spectrocopy signals (both healthy and brain tumour cases) using advanced signal processing algorithms and AI/ML based approaches. The project's tasks include collecting proton MR spectroscopy datasets, develop/implement signal processing algorithms, analysing data and developing a GUI toolbox for lipid suppression. In this research, signal/image processing, MR physics, AI/ML based techniques will be used for medical imaging analysis. MR pulse sequence development will be also be performed to develop/implement hardware based methods for data acquisition and advanced data reconstruction algorithms will also be developed/implemented incorporating lipid suppression.
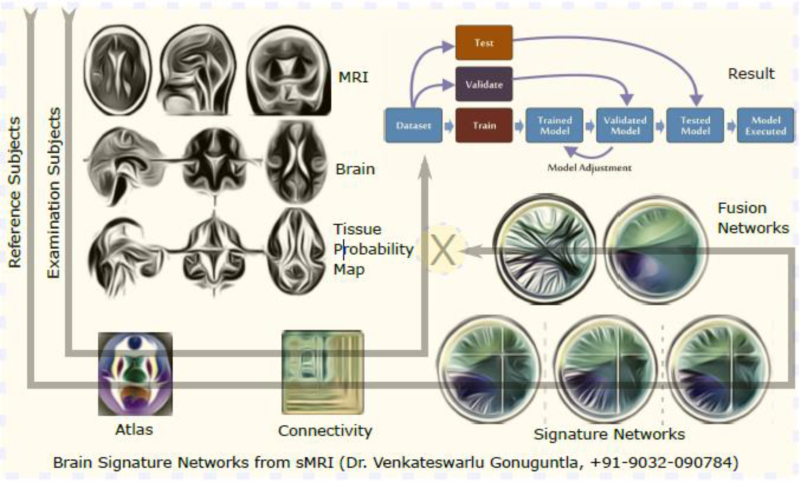
PI: Dr. Venkateswarlu Gonuguntla, Assistant Professor
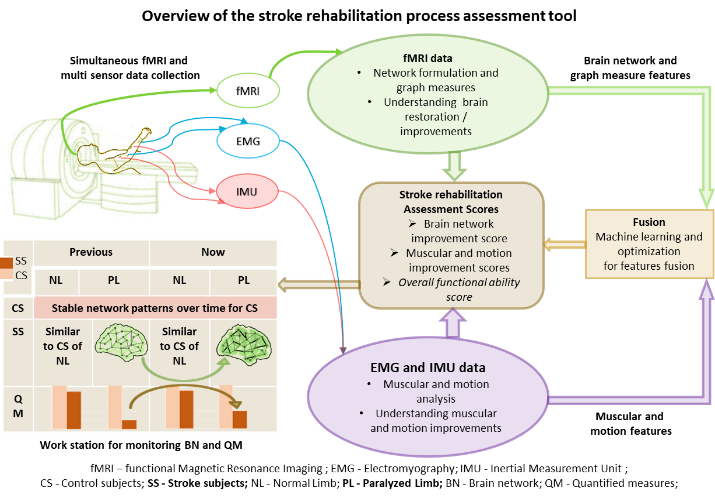
‘The purpose of this study is to integrate a multi-modality system using fMRI, EMG, and IMU into the present rehabilitation and therapy regimens’.
The project's objectives are to:
1. collect fMRI, EMG, and IMU data to monitor the brain, muscles, and movement activities of a stroke patient;
2. develop methods for determining changes in the brain's, muscles', and motion activities at different points; and
3. present an evaluation tool that is simple to integrate into existing treatment plans. This project calls for expertise in machine learning, deep learning, image processing, and signal processing.
PI: Dr. Venkateswarlu Gonuguntla, Assistant Professor
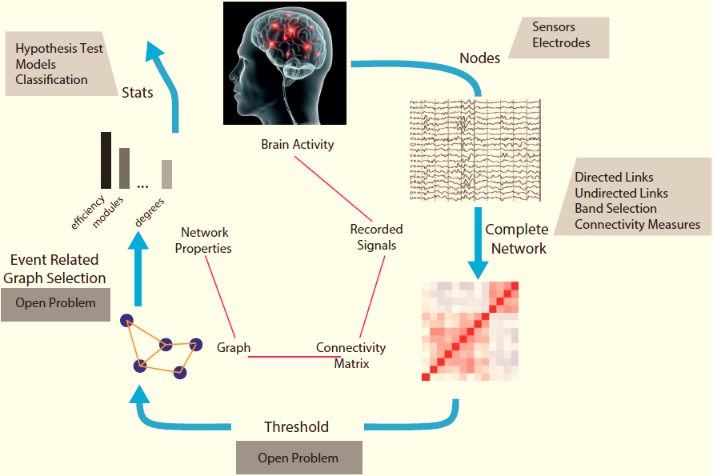
‘The development of EEG-based brain mapping technologies for the analysis and decoding of various neural illnesses will be the main goal of this research’.
The project's tasks include gathering EEG data, signal processing, and interpreting EEG data using network theory and graph theory, as well as using deep learning and machine learning to analyse EEG data. This research makes use of expertise in deep learning, graph theory, machine learning, signal processing, and more.
PI: Dr. Venkateswarlu Gonuguntla, Assistant Professor
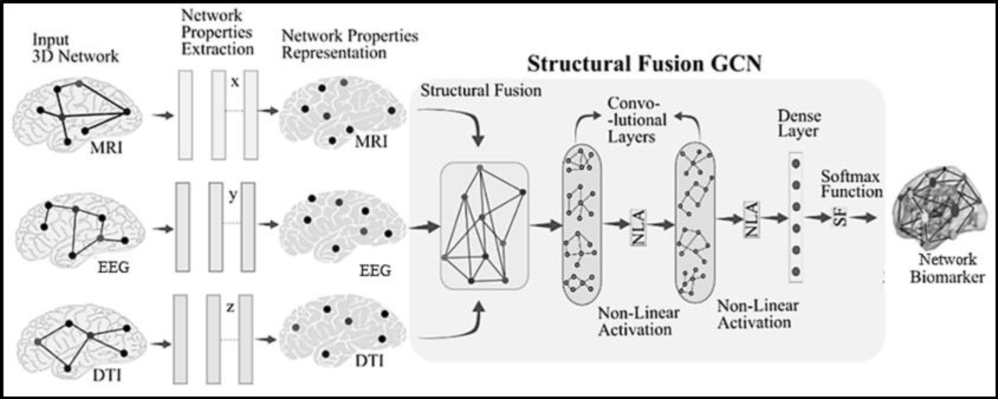
This research develops ‘a. AI-based methods for tracking the development of cognitive impairment and b. MRI structural and functional networks fusion technologies for Alzheimer's disease diagnosis.’
The project's tasks include collecting data from many imaging modalities, analysing data from single and multiple modalities, and developing a GUI for AD early detection. In this research, signal processing and graph neural network techniques are primarily used for medical imaging analysis.
Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: German Sport University, Cologne, Germany
Studying the hip joint loading of various clinical exercises used in the rehabilitation of children with cerebral palsy. The project seeks to evaluate commonly prescribed rehab exercises to investigate their potential to promote optimal bone remodelling and longitudinal growth to minimise femoral lateralisation and hip dysplasia in children with cerebral palsy. The project combines statistical shape modelling and musculoskeletal modelling to create subject specific MSK models which accurately represent the variation in hip geometry seen in children with cerebral palsy.
Copyright: DSHS/Presse und Kommunikation




Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: University of Cape Town, South Africa; SUHRC Anatomy; SUHRC Radiology
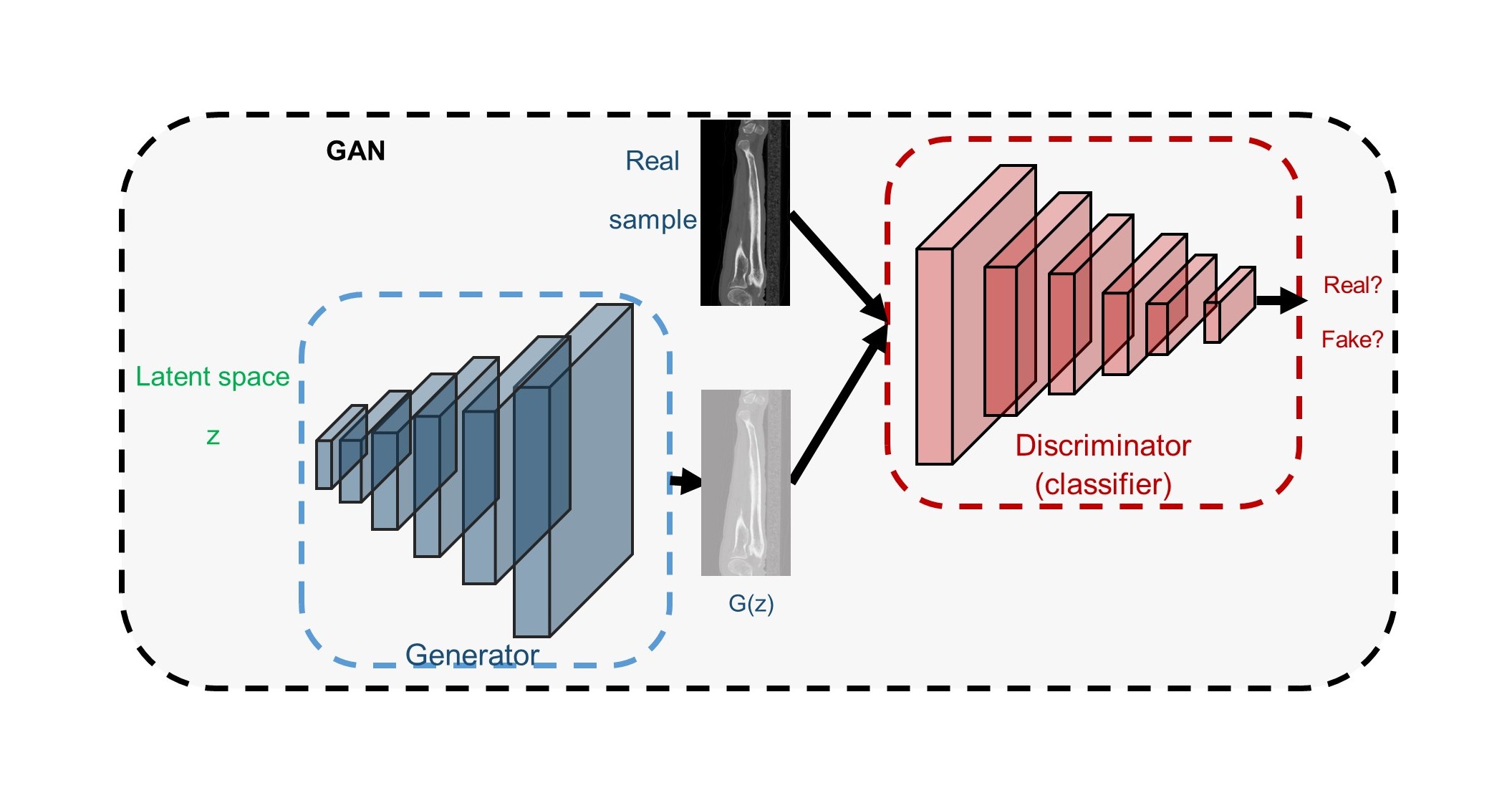
Magnetic resonance imaging (MRI) and computed tomography(CT) are often used for image- based diagnosis of musculoskeletal (MSK) disease and disorders. To leverage complementary information in MRI and CT, and overcome limitations related to either, clinical protocols routinely require both for diagnosis or treatment of MSK disease and disorders. However, image capture from more than one modality results in increased cost to the patient and to the health care system. Furthermore, radiation exposure during CT acquisition limits its use, particularly in immuno-compromised patients or in young children. The synthesis of CT from MRI has become an active research area and particular focus has been on application in radiotherapy..
The aim of this project is to explore two approaches (Atlas -, and VAE - based) for generating sCT from routine MRI images (T1, T2), and evaluate the sCT for use in orthopaedic applications. VAE has not been used for sCT synthesis (a problem defined as mapping two domains) due to the fact it learns mapping within one domain while GAN has shown an ability for multi-domain mapping.
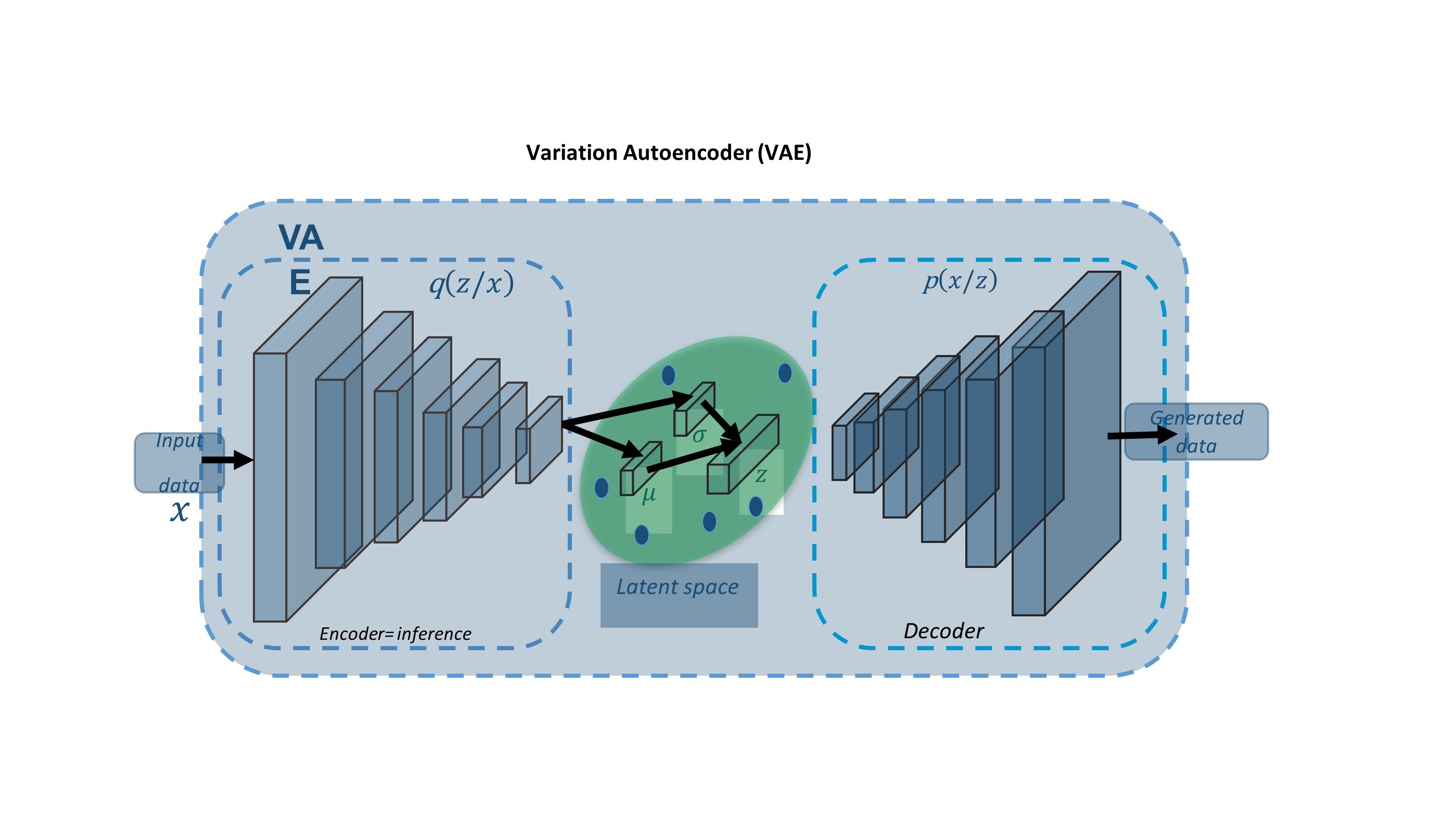
Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: SUHRC Radiology
Osteoarthritis (OA) affects 27 million US adults and often leads to severe disability. The prevalence of OA is 33.6% in adults older than 65 years. Although OA is a widespread and debilitating disease, treatment options are currently limited, and disease modifying therapies have not been established yet. In an effort to develop quantitative biomarkers for OA and to fill the void that exists for diagnosing, monitoring and assessing the extent of early whole joint degeneration in OA, the past decade has shown an increase in using noninvasive imaging for OA. Magnetic Resonance Imaging (MRI) is a central component of large-scale epidemiologic observational studies such as the Osteoarthritis Initiative (OAI), where it can provide a rich array of structural and functional features of musculoskeletal tissues, which in turn shed light on disease etiology, potential treatment pathways, and prognostic tools for long-term disease outcomes. Magnetic resonance (MR)-derived compositional imaging techniques, such as T2 relaxation times, assess the structural and biochemical properties of cartilage since they are sensitive to changes in collagen orientation and water content. Previous studies reported that spatially assessing relaxation times of the knee cartilage using laminar and sub-compartmental analyses could lead to better and possibly earlier identification of cartilage matrix abnormalities..
In this project, we aim to build a fully automated method for the analysis of T2 relaxation time maps with the aim of extracting relevant relaxometry patterns to classify radiographic knee OA in the entire OAI baseline dataset (publicly available). We aim to establish the role of data driven feature extraction to exploit the potential of T2 relaxation times in comparison to classic feature handcrafting. We hypothesize that the coupling of quantitative compositional MRI and deep learning can uncover latent feature representations, non-linear aggregation among elementary features, and thus better characterize OA as compared to compartmental averages or linear patterns decompositions..
Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: IMT Atlantique, Brest, France; SUHRC Radiology
Magnetic resonance imaging (MRI) has emerged as a useful tool for clinicians and scientists to assess the health of cartilage and other soft tissues. Conventional MRI provides sufficient tissue contrast to detect morphological changes in cartilage. However, such changes are detectable only when the cartilage morphology is affected and changes in cartilage physiology prior to morphological changes cannot be visualized or measured with conventional MRI [6]. Detecting these changes through the quantification of physiological, physical, and chemical characterization of the cartilage becomes a priority.
In this project, we aim to address the technical and clinical research gaps by having three objectives as follows: 1) Develop a cartilage specific phantom to be able to acquire multi- scanner data and validate the reliability of the qMRI protocols, 2) develop and validate a non- negative matrix factorization based deep learning framework for the reconstruction of diagnostic quality cartilage relaxation maps from conventional morphologic images, and 3) illustrate the use of the developed phantom and the framework in a cohort with knee OA. The Radiological Society of North America (RSNA) has established the Quantitative Imaging Biomarkers Alliance (QIBA) to address the research gaps by developing qMR imaging guidelines and technical standards documents, referred to as profiles. In our proposal, the phantom development is proposed as per the guidelines of QIBA. No customized phantom exists to date for calibration and multi-site use of cartilage biomarkers. Our proposed phantom development will bridge this gap and will create the necessary ability for conducting multi-center and multi- vendor studies. This will not only establish the reliability of the qMRI protocols, it will also allow us to pool the data together for broader machine learning applications. The second objective of developing deep learning framework will make use of an available repository of Osteoarthritis Initiative imaging dataset. Use of such database will not only allow us to start the algorithm development right from the beginning of the project but will also provide us with the ability to test and explore our abilities for multiple advanced deep learning techniques. We propose to use a non-negative matrix factorization approach to be embedded within the deep learning layer so that we can interpret what our deep learning algorithm is learning. The third objective will allow us to recruit and monitor the OA progression in a borderline OA cohort. This will allow us to prove the efficacy of qMRI protocols and developed phantom. Each of these objectives can be regarded as stand-alone projects that can start simultaneously. Apart from these, the successful completion of the project marks the beginning of new era of qMRI based clinical practice in the management of joint disorders.
Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: University of Cambridge, UK
Femoroacetabular impingement (FAI) is a common cause of hip pain, decreased function, and progression to early osteoarthritis (OA). FAI syndrome is prevalent in young and active adult population such as professional athletes or ballet dancers with impingement exacerbated during the athletic/dance activity. FAI pathomechanics relates to functional impingement due to altered bony anatomy of the acetabulum and proximal femur. FAI can be further categorized as cam-type and pincer-type based on the specific bony deformity present. Impingement due to asphericity of the femoral head and neck is described as cam-type FAI, whereas global or focal over-coverage of the acetabulum results in pincer-type FAI. The most common form of FAI,
However, it is mixed-type impingement where there are features of both cam and pincer morphology. Repetitive abnormal contact between these structures can damage the labrum and articular cartilage if not properly treated and is recognized as a major risk factor in the non- dysplastic hip.
This research project proposes to develop a framework to build personalized treatment strategies and reduce healthcare costs during the management of patients with FAI. The proposal has two aims. The first aim is to develop a medical imaging and modeling framework to provide an in vivo dynamic understanding of FAI using advanced MR imaging techniques and integrate this knowledge in pre-operative planning stage of open/arthroscopic surgery. The second aim is to develop and illustrate the efficacy and validity of medical image synthesis algorithm to create MRI to synthetic CT image volumes and eliminate the need for CT scanning during the open/arthroscopic surgery planning of the individuals with FAI.
Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: University of Missouri-Kansas City, Kansas City, USA
Federated learning avoids the long process of transferring or sharing of datasets from one location (institute) to another. Such data sharing almost always need to have data transfer agreement in place between collaborating institutes. Further, the medical imaging data needs anonymization which is a tricky and time-consuming task. Federated learning is a deep learning technique which allows researchers to train the deep learning algorithms without transferring or sharing data. Data always remain within their respective security walls. This allows data sharing and model training a much easier exercise.
Federated Learning (FL) is simply the decentralized form of Machine Learning. In Machine Learning, we usually train our data that is aggregated from several edge devices like mobile phones, laptops, etc. and is brought together to a centralized server. Machine Learning algorithms, then grab this data and trains itself and finally predicts results for new data generated. Federated Learning is born at the intersection of on-device AI, blockchain, and edge computing/IoT. Here we train a centralized Machine Learning model on decentralized data! The FL architecture in it’s basic form consists of a curator or server that sits at its centre and coordinates the training activities. Clients are mainly edge devices which could run into millions in number. These devices communicate at least twice with the server per training iteration. To start with, they each receive the current global model’s weights from the server, train it on each of their local data to generate updated parameters which are then uploaded back to the server for aggregation. This cycle of communication persists until a pre-set epoch number or an accuracy condition is reached. In the Federated Averaging Algorithm, aggregation simply means an averaging operation. That is all there is to the training of a FL model.
We plan to apply the FL approach in multiple projects where our collaborators are internationally located and are having a huge database. Project specific training and travel may be needed for this project.
Collaborator Institute: University of Cape Town, South Africa; University of Basel, Switzerland; IMT Atlantique, Brest, France
The United Nations and World Economic Forum have estimated that the cumulative loss on the global economy due to non-communicable diseases (NCDs) could reach USD$47 trillion by 2030, should matters remain status quo. Most NCDs perturb the human musculoskeletal system and can have a debilitating impact on quality of life. Musculoskeletal disorders (MSDs) are conditions that can affect muscles, bones and joints, leading to pain and disability. MSDs affect over 30% of the population in Europe and North America leading to larger healthcare expenditure and lost productivity, on top of the negative personal health and social impacts. MSD prevalence data from the global South is by contrast scarce. Medical imaging plays an unequivocal and irreplaceable role in the management of MSDs, with modern imaging systems like X-ray computed-tomography (CT), magnetic resonance imaging (MRI) and three- dimensional (3D) ultrasound (US) providing a variety of methods for capturing the appearance of musculoskeletal structures. While initial emphasis was on diagnostic capacity of medical imaging for MSDs, recent advances have split the medical imaging domain into two – first targeting image acquisition techniques aimed to improve image resolution and tissue contrast and second targeting image post-processing techniques to derive cost-effective, efficient and reliable methods for processing and efficiently analysing the imaged data. It is this sub-domain of image post-processing which has seen the confluence of advances from computer science, including machine learning, big data analytics, and artificial intelligence.
This project proposes to build statistical inference model(s) of the entire human musculoskeletal structure with the express intention of infusing a critical rethinking of clinical practices in the management of MSDs. We call this a “virtual statistical multi-domain, multi- scale musculoskeletal human - VSM3-HUMAN” modelling framework.
This project adheres to a holistic approach at three levels: 1) the entire musculoskeletal system will be modelled as an integrated system, 2) all collected image data will be used in the analysis, and 3) the approach will allow a switch between a global and local perspective on-demand while retaining all patient information in the analysis. Further, the proposed technology pushes beyond state-of-the-art since the holistic approach for data-driven modelling will demand addressing of new challenges. These include, but are not limited to, 1) developing the models beyond state-of-the-art limitation of two or three organs, 2) developing a framework to retain statistical correlation in the entire musculoskeletal structure, 3) developing new methods that go beyond traditional correspondence based statistical modelling, 4) generalizing statistical modelling from linear principal component analysis (PCA) based spaces to non-linear spaces, 5) incorporation of multimodal (MRI and CT) image information in the same model, and 6) contention with new standards in model validation.
Pl : Dr. Bhushan Borotikar, Associate Professor
Collaborator Institute: SUHRC Nutrition
Obesity has become a global pandemic with multiple adverse clinical consequences. Accumulating evidence demonstrates that cognition is affected by an excess of body adiposity in both adults and children. Epidemiological studies also indicate that midlife obesity increases the risk of progression to mild cognitive impairment (MCI) and Alzheimer's disease (AD). On the contrary, higher body mass index (BMI) in late-life might be protective.8 This obesity paradox might be associated with the confounding effect of weight loss in preclinical AD. The exact mechanisms leading to cognitive impairment and neurodegeneration in persons with an excess of body adiposity remain to be fully elucidated. Animal models suggest a significant contribution of obesity and obesity-related metabolic disturbances to AD pathophysiology. In contrast, human studies assessing the impact of obesity on amyloid and tau pathology report conflicting findings both in vivo and in post-mortem studies. Thus, higher BMI has been related to higher, but also to lower, AD burden. On the other hand, obesity might contribute to neurodegeneration by mechanisms unrelated to AD. Obesity is a state of peripheral low-grade chronic inflammation, and it is frequently associated with an abnormal peripheral sensitivity to insulin effects. In experimental models, obesity-related peripheral inflammation has been linked to blood-brain barrier dysfunction, neuroinflammation, and neurodegeneration, whereas central insulin resistance has been associated with impaired synaptic plasticity and memory. Furthermore, obesity is a strong risk factor for hypertension, type 2 diabetes, and dyslipidemia, and it is a well-established cerebrovascular risk factor.
The proposed project is a new project and we aim to collect quantitative neuroimaging data on obese and healthy population to study the inflammatory and cognitive markers of obesity.
Collaborator Institute: University of Missouri-Columbia, USA
Bone age in the pediatric and adolescent population is a crucial and highly sought-after metric. The progress of physical maturation of young individuals can be used as a biological marker connected to aging. Estimating biological age from physical development is therefore a highly important topic in both clinical and legal (forensic) medicine applications. In clinical medicine, biological age estimation is motivated by the diagnosis of endocrinological diseases like accelerated or delayed development in adolescents, or for optimally planning the time-point of pediatric orthopedic surgery interventions while bones are still growing. Examples of such interventions include leg-length discrepancy correction or spinal deformity correction surgery. In legal medicine, when identification documents of children or adolescents are missing, as may be the case in asylum-seeking procedures or in criminal investigations, estimation of physical maturation is used as an approximation to assess unknown chronological age. However, many propose that the non-medical results are inconclusive and thus medical bone age estimation is necessary in such cases.
X-rays have been frequently used for image acquisition as they are a fast and inexpensive method of examination. But the ionizing radiation exposure is considered “harmful to the body” and should be used only as a last choice in the pediatric population after weighing in the associated risk. Besides, the image examination to determine bone age is a tedious task and is subjective to the radiologist or expert conducting the analysis. A major drawback of the widely used biological (bone) age as estimated by the radiology approach is exposure to ionizing radiation, which cannot be justified in legal medicine applications for examining healthy children and adolescents without any diagnostic reasons. Over the years, research in age estimation has exhibited a tremendous interest in magnetic resonance imaging (MRI) as an ionizing radiation-free alternative to develop new schemes for age estimation. The availability of big data and high computational performance has led to the use of Artificial Intelligence in almost every domain including medical imaging. Recently, the use of deep convolutional neural networks (DCNN) has shown to be immensely successful in solving diverse machine learning and computer vision problems, mainly due to their ability to automatically learn task-relevant features from large training datasets.
It is observed that most of the studies in the literature are focused on imaging data of hand scans. In the case of clinical medicine application, when a patient’s other anatomical site scans are already available, then to identify bone age to confirm the course of treatment through surgeries, they are further exposed to unnecessary ionizing radiations on the hands. Thus, to eliminate this exposure, deep learning framework can be effectively used for automated bone age estimation on various anatomical sites using different medical imaging data. This project focuses on knee MRI on pediatric datasets.
Collaborator Institute: SUHRC Radiology, USA
The project could be seen as an extension of MRI to CT synthesis, but has multiple new paradigms in terms of paired data availability, new data generation/augmentation techniques, data validation techniques, 3-way data registration techniques, multi-domain adaptation technique, model generalizability demonstration, model direction flexibility (MRI to CT, CT to X- ray, X-ray to MRI etc.). More details could be discussed with me.